Revolutionary Low-Cost Phenotyping System Reveals Key Insights for Breeding Disease-Resistant Crops!
2024-11-20
Author: Jacques
Introduction
A groundbreaking research study has unveiled a cost-effective phenotyping approach that provides essential insights into the genetic diversity of Quantitative Disease Resistance (QDR) in wild tomatoes. This innovative system not only aids in the understanding of plant resilience but also opens new avenues for breeding disease-resistant crops, potentially transforming agricultural practices.
Understanding QDR
QDR represents a sophisticated and durable form of resistance that gives plants partial protection against a wide array of pathogens, unlike traditional qualitative resistance, which relies on major resistance (R) genes. QDR is polygenic, featuring various protective mechanisms, including delayed lesion development and reduced infection rates. The complexities surrounding the genetic and regulatory aspects of QDR have often posed significant challenges, primarily due to the need for advanced technology and in-depth data analysis.
The Study
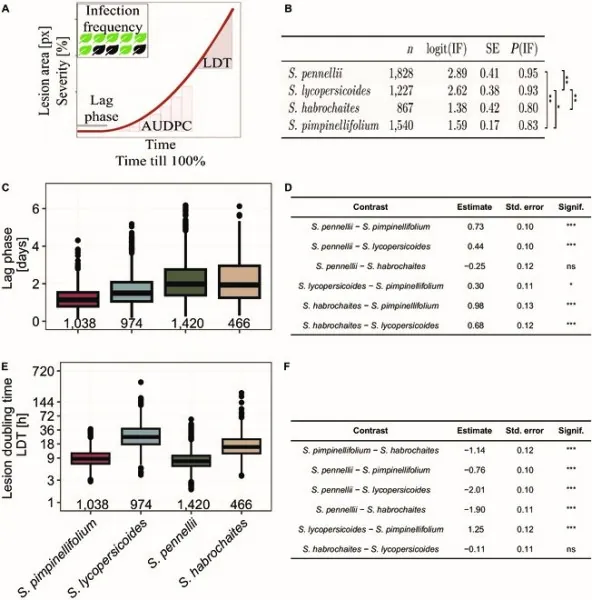
In a pivotal study published in Plant Phenomics on August 5, 2024, researchers employed the "Navautron" automated phenotyping system to investigate QDR in four wild tomato species: S. habrochaites, S. lycopersicoides, S. pennellii, and S. pimpinellifolium, examining their resistance against the pathogen Sclerotinia sclerotiorum. This advanced phenotyping system captured continuous imagery of infected leaves and quantified crucial parameters, including Infection Frequency (IF), Lag-Phase Duration (LPD), Lesion Doubling Time (LDT), and the Area Under the Disease Progress Curve (AUDPC).
Key Findings
The results were illuminating! The study discovered significant phenotypic diversity in QDR across the tested species. Notably, S. pimpinellifolium showcased the shortest lag phase at just 36.2 hours, while both S. habrochaites and S. pennellii had longer phases nearing 59 hours. Additionally, lesion growth rates differed dramatically; S. pimpinellifolium and S. pennellii displayed rapid lesion expansion with doubling times of only 11 hours, compared to S. habrochaites and S. lycopersicoides, which expanded much slower, spanning up to 41 hours.
Intraspecific Variations
Notably, the study assessed intraspecific variations, revealing a diverse range of disease response among accessions of S. pennellii, whereas S. lycopersicoides displayed greater consistency in resistance. Particular attention was given to the accessions of S. pennellii, showing genetic variation with LA1941 demonstrating the most resistance and LA1809 exhibiting the highest susceptibility.
Implications and Future Directions
Dr. Remco Stam, the lead researcher, highlighted the implications of these findings, stating, “Unlocking the potential of QDR in crop breeding has been a long-standing challenge. Our study showcases a cost-effective phenotyping system that can provide high-resolution data crucial for understanding and utilizing QDR traits in wild crop relatives.”
Conclusion
The need for sustainable agriculture is more pressing than ever, and this research reinforces the value of low-cost, high-efficiency phenotyping in plant pathology. With the ability to break down complex QDR into manageable components, scientists are optimistic about breeding resilient crops that can thrive amid environmental stresses and disease threats—without heavy reliance on chemical fungicides or the limitations of conventional R-genes.
This study not only paves the way for advancing agricultural practices but also brings hope for the future of food security, advocating for a shift towards sustainable and resilient crop varieties. Stay tuned as the field of plant breeding continues to evolve!



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)