Revolutionary Tool Maps Protein Interactions to Combat Lung Cancer!
2024-11-21
Author: Benjamin
Revolutionary Tool Maps Protein Interactions to Combat Lung Cancer!
In an exciting breakthrough that could reshape the approach to treating lung cancer, a team of researchers led by Ahmet F. Coskun is making strides in their mission to create a comprehensive 3D atlas of the human body. Their latest findings focus on mapping protein interactions—an essential factor in understanding and potentially overcoming the challenges of non-small cell lung cancer (NSCLC).
Innovative Research and Technology
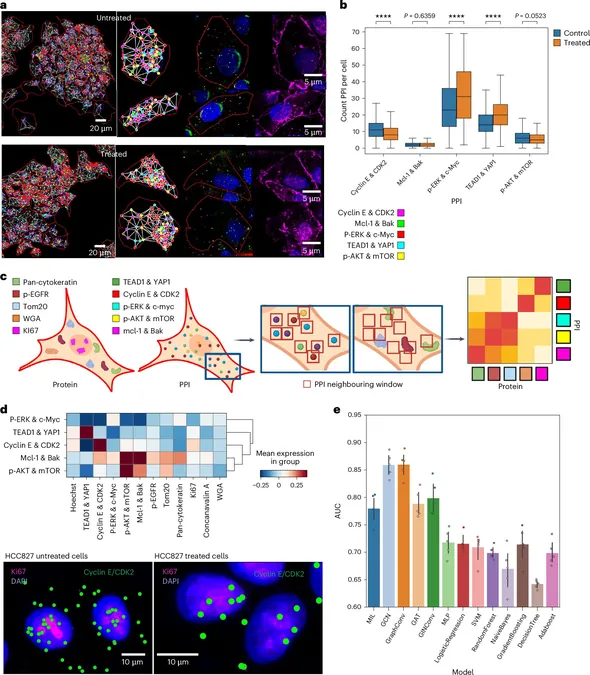
Coskun and his team are at the forefront of pioneering a cutting-edge field known as "spatial interactomics." This discipline investigates how various biomolecules interact within individual cells, facilitating deeper insights into cancer biology and treatment resistance. Their innovative technique, dubbed the "intelligent sequential proximity ligation assay" (iseqPLA), is revolutionizing the way scientists can visualize these critical interactions at the subcellular level.
Understanding Protein Interactions
"Think of iseqPLA as a sophisticated test that illuminates protein interactions in space," explains Coskun, who holds a prestigious position as the Bernie Marcus Early Career Professor in the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University. "For the first time, we’re creating a new area of research that reveals how cells physically communicate and modulate their behavior through protein interactions."
Tackling Drug Resistance
One of the immediate objectives of this research is to uncover how these protein-protein interactions contribute to drug resistance in NSCLC. With existing therapies such as tyrosine kinase inhibitors (TKIs) like Osimertinib proving effective for a time, many patients ultimately develop resistance—a frustrating hurdle in treatment. The Coskun team is determined to investigate the molecular interactions that may underlie this resistance.
Efficiency and Advancements
Rather than analyzing only a handful of interactions at a time, iseqPLA allows researchers to examine up to 47 protein interactions in a single sample. This efficiency streamlines the research process, enabling faster and more resource-effective studies. Coskun likens iseqPLA to creating a "super detailed map" illustrating how proteins within a cell connect and interact.
Leveraging Data with Computer Modeling
Additionally, to enhance their findings, the team has developed a computer model to analyze the abundant data generated by iseqPLA, identifying patterns in protein interactions that can help predict how individual cells respond to treatments. "We’ve shown that iseqPLA is effective not just in cultured cells but also in tissue samples from both mice and humans," Coskun highlights.
Future Aspirations and Broader Applications
Looking ahead, Coskun envisions expanding the capabilities of iseqPLA to explore interactions not just among proteins, but also between RNA, metabolites, and other subcellular components. He plans to establish a startup, named SpatAllize, to bring this groundbreaking technology to market, with the support of VentureLab at Georgia Tech.
Broader Implications for Health
But the ambitions don’t stop there! Coskun intends to apply iseqPLA in other critical areas of research, analyzing how protein interactions can influence immune responses and the health of the heart and brain. His forward-thinking approach includes integrating iseqPLA with advanced imaging technologies and automated deep learning, ultimately paving the way for robotics in spatial interactomics.
Revolutionizing Cancer Research
"This will enable us to map all molecules within cells and tissues, significantly enhancing our understanding of drug-cell interactions, particularly for personalized cancer treatment plans," asserts Coskun.
Stay tuned as Coskun and his team continue on their quest to revolutionize cancer research and treatment through the power of spatial interactomics. With each breakthrough, they are not only opening new doors for understanding disease mechanisms but also paving the path toward more effective therapies for patients in need.








 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)