Unveiling the Secret Weapon of a Giant Virus: A Breakthrough in Protein-Making Machinery!
2024-12-18
Author: Sophie
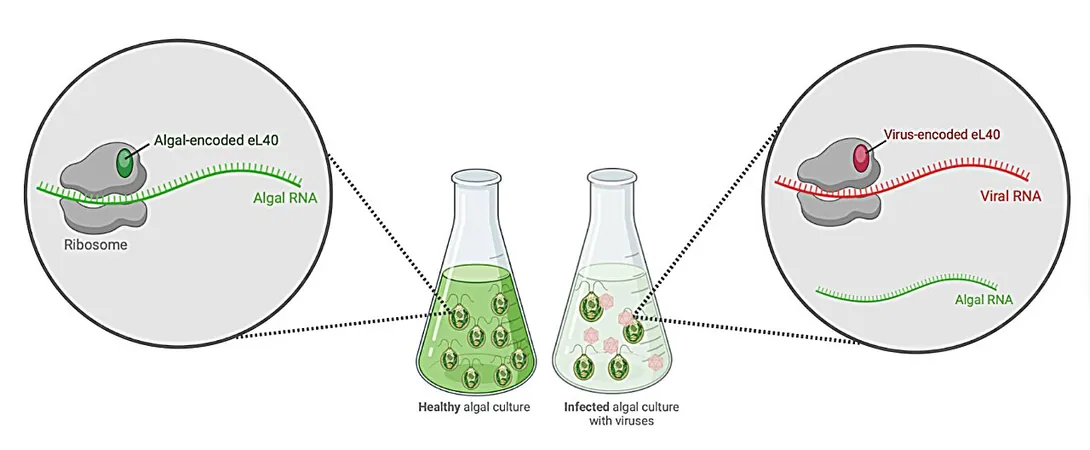
In a groundbreaking study, researchers from the University of Hawai'i at Mānoa have unveiled a fascinating aspect of virology: the FloV-SA2 virus surprisingly encodes a crucial protein essential for the formation of ribosomes. These ribosomes are considered the powerhouses of all cells, responsible for translating genetic code into proteins, which in turn serve as the building blocks of life itself. This monumental discovery marks the first instance of a eukaryotic virus—one that infects higher organisms like plants and animals—demonstrating the ability to encode such a vital protein.
This pivotal research has been published in the prestigious journal *npj Viruses* and has the potential to transform our understanding of how viruses interact with their host organisms.
The Complexity of Viruses: More than Just Pests
Viruses, often regarded as mere genetic material wrapped in a protein shell, possess intricate mechanisms for replication. While simpler viruses rely predominantly on their host cells' machinery, larger, "giant" viruses like FloV-SA2 have evolved to create several proteins that facilitate their own reproduction. Julie Thomy, a leading researcher on the project and postdoctoral scholar at the Daniel K. Inouye Center for Microbial Oceanography, expressed her enthusiasm about this discovery, stating, “Finding that this virus encodes a ribosomal protein named eL40 is both exciting and unexpected.”
Thomy added that while the concept of a virus modifying critical cellular machinery was plausible, there had never been evidence of it occurring in a eukaryotic virus—until now.
A Deep Dive into the Discovery
The FloV-SA2 virus was isolated as part of an extensive endeavor by the Marine Viral Ecology Laboratories (MarVEL) at SOEST, aimed at exploring the diverse viral inhabitants of our oceans. Christopher Schvarcz, a former Oceanography graduate student, played a vital role by collecting water samples from Station ALOHA, situated 60 miles off the coast of O'ahu, where he successfully isolated a plethora of viruses, including the intriguing FloV-SA2, which specifically targets a phytoplankton species known as Florenciella.
“Chris’s remarkable productivity led to an overwhelming number of samples, and it took time for us to analyze them thoroughly,” remarked Grieg Steward, the faculty member overseeing the project. “The subsequent analysis conducted by Dr. Thomy was certainly worth the wait.”
The Intriguing Question of Virus Protein Preference
Past investigations have revealed that giant viruses, like FloV-SA2, possess genes that code for proteins linked to various metabolic processes. Some of these functions, such as fermentative processes or light detection, have raised eyebrows among scientists. Presently, the researchers are on a quest to discern how this ribosomal protein functions in the context of the virus.
"Our leading hypothesis posits that by integrating its own protein into the ribosome, the virus might modify this essential machinery to prioritize the production of viral proteins over standard cellular proteins," Thomy explained.
The Bigger Picture: Viruses in Marine Ecosystems
Viruses are more than mere nuisances; they play a significant role in the stability of ocean ecosystems. Steward emphasized their influence on biological productivity, community dynamics, and evolutionary transformations. This discovery sheds light on the intricate interplay between viruses and phytoplankton—the foundational components of marine ecosystems—while paving the way for future research into viral biology's fundamental principles.
Expectations are high that the FloV-SA2 virus will serve as an invaluable model for probing into the sophisticated strategies viruses employ to manipulate host metabolism, allowing them to commandeer cellular resources and energy to flourish.
Stay tuned as this exciting field of research unfolds, potentially unlocking the mysteries of viral behavior and their profound impacts on the environment.









 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)