Battle of the Biopsies: Which Single-Cell Profiling Method Reigns Supreme for Gut Studies?
2025-04-15
Author: Sarah
In the relentless quest to decode gastrointestinal diseases—especially the notorious cancer—research has predominantly zeroed in on epithelial cells. These vital cells line our organs and play crucial roles, yet they can be tricky to study. The reason? Their delicate nature makes them prone to damage during the challenging extraction process from the gut's mucosal lining.
Enter the two heavyweights in the ring: Droplet-based single-cell RNA sequencing (D-scRNA) and the newer Picowell-based platforms (P-scRNA). While D-scRNA has been a powerhouse in providing insights, it comes at a cost—it's resource-intensive and can stress those vulnerable epithelial cells. Meanwhile, P-scRNA touts a gentler, more efficient approach, but the question looms: How do they stack up against one another in real-world human gut biopsies?
A groundbreaking study published in *Cellular and Molecular Gastroenterology and Hepatology* sought to answer this burning question by comparing D-scRNA and P-scRNA head-to-head.
The findings reveal a complex landscape with unique pros and cons for each method. These differences pose significant hurdles when attempting to achieve consistency and reproducibility across different research studies.
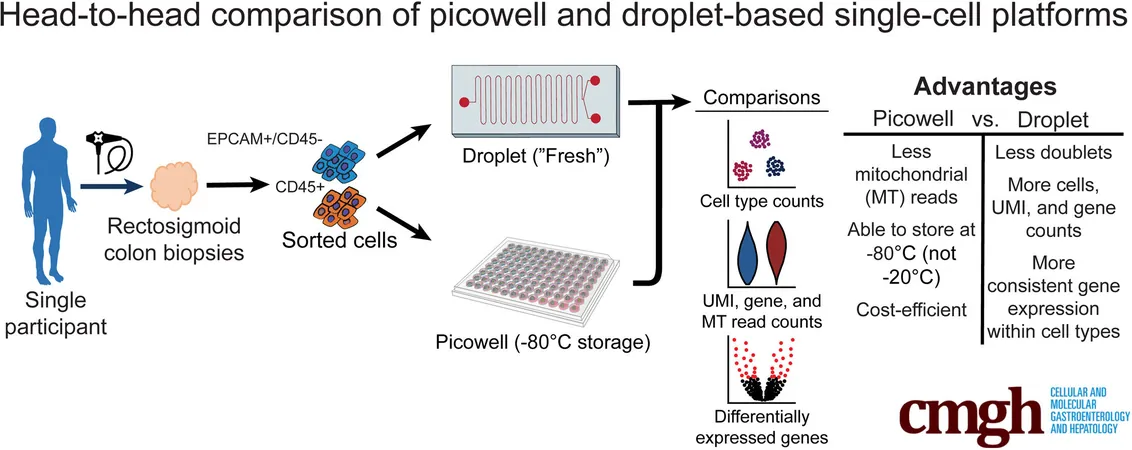
To ensure clarity in their comparison, researchers meticulously collected human colon biopsies using identical protocols, enabling a direct analysis of the same cellular population through both D-scRNA and P-scRNA. They scrutinized numerous factors like cell count, gene coverage, and mitochondrial DNA contamination.
The results were illuminating. D-scRNA emerged as a champion for depth, capturing a greater number of cells with detailed gene profiles. However, this advantage comes with caveats, including considerable resource demands and a higher likelihood of mitochondrial contamination, potentially skewing results.
On the flip side, P-scRNA proved to be more cell-friendly and efficient, better preserving cell quality and reducing mitochondrial contamination. Yet, its capabilities fell short when it came to capturing a diverse array of cell types—especially the rarer ones that might hold the key to understanding specific diseases.
So, what does this mean for researchers? The insights gained from this study can guide scientists in interpreting their scRNA data and choosing the right method for their specific investigations, saving time and resources in the process. By sharing these findings, the research community can navigate the complexities of single-cell technologies, ultimately accelerating breakthroughs in gastrointestinal health.




 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)