Boltz-1 Launch: An Open-Source Revolution in Biomolecular Structure Prediction
2024-12-17
Author: Mei
In a groundbreaking move for biomedical research, MIT scientists have unveiled Boltz-1, a fully open-source AI model poised to compete with Google DeepMind's AlphaFold3 in predicting the intricate 3D structures of proteins and other biomolecules. This revolutionary model aims to propel advancements in drug development and biomedical discovery, making it a game-changer in the field. The detailed research paper is available on the bioRxiv preprint server.
Boltz-1 emerges from the MIT Jameel Clinic for Machine Learning in Health, where graduate students Jeremy Wohlwend and Gabriele Corso spearheaded its development along with leading researchers, including Saro Passaro and MIT professors Regina Barzilay and Tommi Jaakkola. During a recent event at MIT's Stata Center, Corso expressed the ambition behind Boltz-1, stating, "We hope for this to be a starting point for the community," emphasizing the model's open nature and the desire for collaboration.
Understanding a protein's structure is crucial because it directly influences its function. The ability to accurately predict these structures is vital for the design of new drugs and engineered proteins with specific functions. Traditionally, this has been a daunting challenge due to the complexity involved in folding amino acid chains into their corresponding 3D structures.
AlphaFold2 laid the groundwork for addressing this issue, receiving widespread acclaim—including the 2024 Nobel Prize in Chemistry for its developers. This model utilizes cutting-edge machine learning techniques to generate 3D protein structures that rival those derived from laboratory experiments. Building on AlphaFold2's success, AlphaFold3 has refined the process by incorporating a generative diffusion model to manage the intrinsic uncertainty in predicting complex structures. However, AlphaFold3 remains proprietary and unavailable for commercial use, prompting a call for more accessible alternatives.
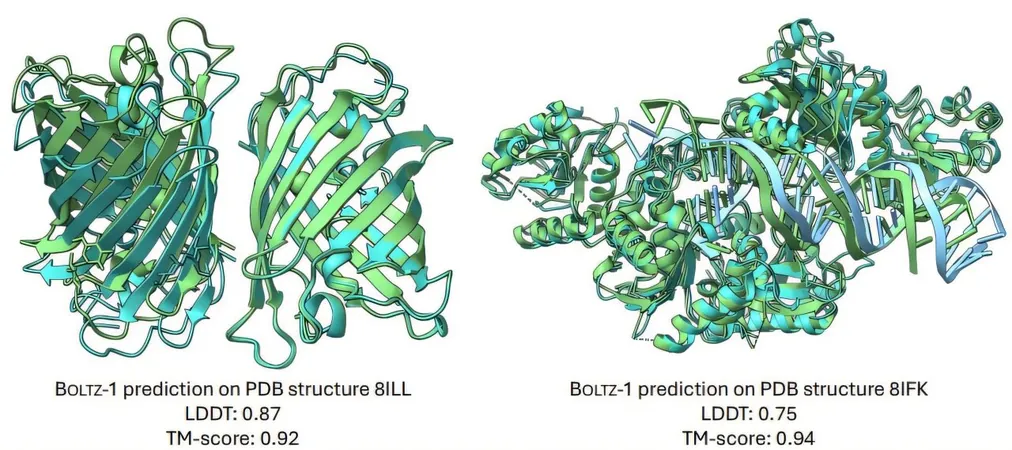
In contrast, Boltz-1 not only matches the accuracy of AlphaFold3 across a diverse range of biomolecular predictions but also opens up its architecture and training processes for public use. This ensures that researchers globally can adapt and enhance the model, thus fostering broader scientific collaboration.
Barzilay commended the team's dedication, noting, "There are many exciting ideas for further improvements, and we look forward to sharing them in the coming months." The creation of Boltz-1 involved months of research, with the team facing significant hurdles in managing the inconsistencies found in the Protein Data Bank—a comprehensive repository of biomolecular structures.
Experts are already heralding Boltz-1 as a significant milestone. Mathai Mammen, CEO of Parabilis Medicines, described it as a "breakthrough" model that opens new avenues in structural biology, stating, "This landmark effort will accelerate the creation of life-changing medicines." Similarly, Jonathan Weissman, an MIT biology professor, emphasized the model's potential to catalyze a wave of discoveries across the scientific community.
The future looks bright for Boltz-1, with the MIT team committed to enhancing its performance and reducing prediction times. They invite researchers to experiment with Boltz-1 via their GitHub repository and engage with fellow users through collaborative platforms.
As Daniele De Rossi, a prominent biochemist, aptly put it, "Boltz-1's open-source nature signifies a new dawn for biomolecular modeling—a tool that empowers every researcher, accelerates discovery, and redefines the future of medicine." The ripple effect of this innovation could lead to unprecedented advancements in biomedical research, making Boltz-1 a pivotal resource for scientists everywhere.



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)