Groundbreaking Study Reveals Surprising Role of Antibiotic Resistance Genes in Gut Microbe Adaptability
2025-04-07
Author: Rajesh
Groundbreaking Study Reveals Surprising Role of Antibiotic Resistance Genes in Gut Microbe Adaptability
A new study from researchers at the Institute of Science and Technology Austria (ISTA) has unveiled unexpected insights into the role of antibiotic resistance genes in gut microbes. Instead of merely serving as mechanisms for discarding antibiotics, these genetic networks may play a vital role in helping bacteria adapt and thrive in their often tumultuous environments.
Published in the Proceedings of the National Academy of Sciences, this research centers on the intricate "mar" gene regulatory network, which has long been associated with multiple antibiotic resistance. Surprisingly, it was discovered that this network maintains a low level of genetic activity even when it is supposedly dormant, a phenomenon that researchers believe is key to the bacteria's ability to adjust to the rapidly changing conditions of the gut.
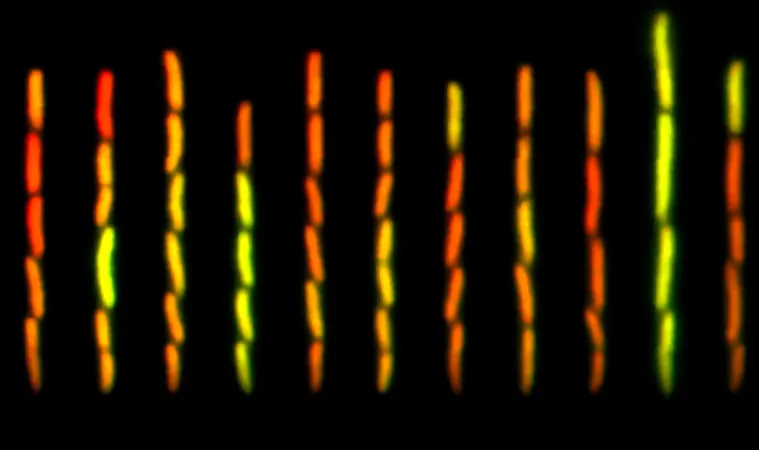
Lead researcher Kirti Jain, alongside Professor Calin Guet, highlighted a unique aspect of the mar system—the presence of pulsatile gene expression, where signals pulse in sync with the host's feeding patterns. This mechanism enables gut microbes to effectively manage their growth cycles in response to nutrient availability. “We do not know of another mechanism that got selected in the OFF state,” Guet remarked, referring to the adaptive advantage conferred by this unexpected genetic behavior.
The mar system, particularly well-studied in E. coli, illustrates a paradox. It remains tightly regulated to manage antibiotic resistance yet allows for some functional expression during its "OFF" state. Jain posed the critical question: “If the mar system evolved under strong selective pressure to be tightly controlled, why does it still permit low-level basal expression?” This inquiry prompted the research team to explore the evolutionary implications of this basal gene activity.
In a fascinating twist, the researchers discovered that the mar system is initiated by a rare transcription start signal known as the “start codon,” which has a peculiar GTG sequence rarely found in bacterial DNA. By mutating this codon, the team observed significant changes in the network's expression levels. Importantly, the wild-type bacteria maintained expression patterns that aligned with their host's feeding schedules, enabling optimal resource usage in an unpredictable environment.
The study also sheds light on the auxiliary role of antibiotic resistance mechanisms, suggesting that these "pumps" which expel antibiotics might have evolved as a means to flush out not just antibiotics but also various toxins introduced through the host's diet. Maintaining such mechanisms is energetically costly for microbes, indicating that the primary function of the mar system likely extends beyond simple resistance, emphasizing their adaptability to survive amid competing strains of bacteria.
This research holds potential implications for public health, as understanding these bacterial behaviors could pave the way for innovative therapeutic strategies that target the broader adaptability of gut microbes rather than merely their resistance to antibiotics. Jain emphasized the importance of questioning established assumptions in light of technological advancements, noting the value of interdisciplinary collaboration that shaped their analysis.
In conclusion, these findings not only challenge existing beliefs about antibiotic resistive gene networks but also hint at the remarkable adaptability of gut bacteria, an insight that could redefine future approaches to tackling antibiotic resistance in healthcare settings.





 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)