Revolutionary Low-Cost Phenotyping System Sheds Light on Disease Resistance in Wild Tomatoes!
2024-11-20
Author: Siti
Introduction
In an exciting breakthrough for agricultural research, scientists have unveiled the potential of quantitative disease resistance (QDR) in wild tomatoes—a pivotal discovery that could reshape the future of crop resilience against pathogens. Unlike qualitative resistance, which relies on major resistance genes, QDR is polygenic and offers a more durable form of protection, manifesting in ways such as delayed lesion development and reduced infection frequency.
Study Overview
A recent study published in Plant Phenomics on August 5, 2024, reveals the untapped potential of wild tomatoes in breeding new, disease-resistant varieties. The researchers focused on four wild tomato species: Solanum habrochaites, Solanum lycopersicoides, Solanum pennellii, and Solanum pimpinellifolium, assessing their resistance against the notorious pathogen Sclerotinia sclerotiorum using the innovative "Navautron" automated phenotyping system.
Methodology
This cutting-edge technology continuously captured high-resolution images of infected leaves, utilizing advanced algorithms to measure key indicators such as infection frequency (IF), lag-phase duration, lesion doubling time (LDT), and the area under the disease progress curve (AUDPC). By employing statistical models like generalized least squares and generalized linear models, the research team found significant phenotypic diversity in QDR among the wild tomato species.
Findings
Notably, S. pimpinellifolium exhibited the shortest lag phase at just 36.2 hours, demonstrating its potential as a valuable genetic resource for breeding programs. On the contrary, S. habrochaites and S. pennellii displayed longer lag phases, around 59 hours, indicating varying levels of resistance.
The analysis of lesion growth rates revealed that S. pimpinellifolium and S. pennellii experienced the fastest lesion expansion, with doubling times of 11 hours, while S. habrochaites and S. lycopersicoides exhibited significantly slower rates—maxing out at 36 and 41 hours, respectively. The infection frequency varied across species, with S. habrochaites reporting the lowest (80%), while S. lycopersicoides and S. pennellii had higher frequencies at 93%-95%.
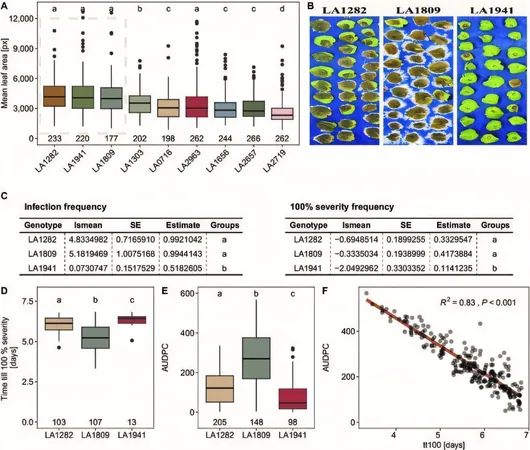
Intraspecific Variation
In a deeper dive into intraspecific variation, S. pennellii showed a wide range of lag phases across different accessions. For instance, accession LA1941 stood out with the best resistance profile, demonstrating the potential for targeted breeding strategies based on genetic insights. In contrast, LA1809 recorded the highest severity of infection, underscoring the significance of genotype-dependent resistance.
Conclusion
Dr. Remco Stam, leading the research, emphasized the importance of these findings, stating, "Unlocking the potential of QDR in crop breeding has been a long-standing challenge. Our study highlights a cost-effective phenotyping system that can provide high-resolution data crucial for understanding and harnessing QDR traits in wild crop relatives."
This revolutionary low-cost phenotyping system not only paves the way for future studies in plant resilience but also holds promise for global food security, addressing the critical challenge of crop loss due to diseases. The potential to breed more resilient tomato varieties signals hope for farmers confronting increasingly aggressive plant pathogens, ultimately leading to healthier crops and improved agricultural sustainability.

 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)