Revolutionary Nanopore Technology Unlocks Secrets of Long Protein Strands!
2024-11-27
Author: Sarah
Revolutionary Nanopore Technology Unlocks Secrets of Long Protein Strands!
Recent advancements in Oxford Nanopore technology are paving the way for unprecedented discoveries in molecular biology. This groundbreaking method has previously been utilized on the International Space Station to unravel genomic changes linked to the harsh conditions of spaceflight. Its applications extend far beyond the cosmos—this technology has played a vital role in identifying disease origins in remote locations, analyzing extremophiles in Antarctica, and investigating food contamination sources.
But why is this innovation crucial now more than ever? As humanity prepares for potential exploration beyond our planet, advanced sequencing technologies like nanopore reading are becoming essential tools for astronauts and scientists alike.
The latest breakthrough involves the ability to sequence single protein molecules in their native, full-length form, offering a more profound insights into proteomic diversity. Unfortunately, previous technologies have struggled to achieve this level of precision.
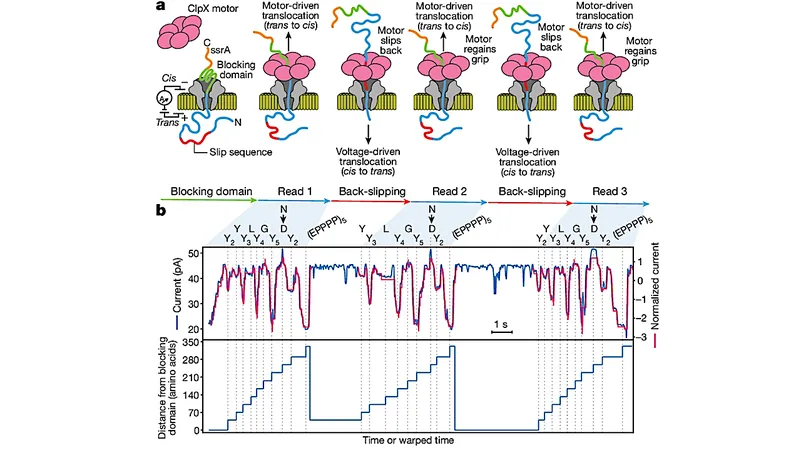
In a significant advancement, researchers have introduced a novel technique for long-range, single-molecule reading of intact protein strands utilizing a commercial nanopore sensor array. This method employs the ClpX unfoldase to guide proteins through a CsgG nanopore, revealing that ClpX translocates these proteins in two-residue steps—a discovery that holds immense potential for biochemistry.
This innovative approach enables researchers to detect single amino acids within synthetic protein strands, which can be hundreds of amino acids long. This makes it possible to locate specific amino acid substitutions and to analyze post-translational modifications like phosphorylation—changes that can dramatically affect a protein's function.
Moreover, the technology allows for multiple readings of individual protein molecules, enhancing classification accuracy and opening doors to precise protein barcode sequencing. This could revolutionize the field, offering a reliable method for characterizing proteins at an individual level.
In a further complication to the traditional understanding, researchers have developed a biophysical model that simulates raw nanopore signals before the experiment, based on residue volume and charge. This advancement enhances the interpretation of raw signal data, leading to clearer, more actionable insights.
The methods developed have been applied to analyze full-length, folded protein domains, providing a complete end-to-end analysis that could redefine our understanding of protein structure and function. This proof of concept ushers in a new era of proteomics, allowing scientists to identify and characterize full-length proteoforms at single-molecule resolution.
Stay tuned as the future unfolds—this technology may soon provide answers about the complex roles proteins play in human health, disease, and potentially even our survival as we venture into new worlds!


 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)