Revolutionary Sludge Sequencing Unlocks Secrets of Antibiotic Resistance in Wastewater!
2024-10-09
Author: Daniel
Introduction
In a groundbreaking study, researchers have unveiled the extraordinary complexities of microbial life hidden within activated sludge used in wastewater treatment. These microbial communities play a pivotal role in eliminating pollutants, but traditional methods often overlook the intricate web of interactions they support. To address this gap, scientists turned to high-throughput single-cell sequencing techniques that promise to revolutionize our understanding of these essential ecosystems.
Research Overview
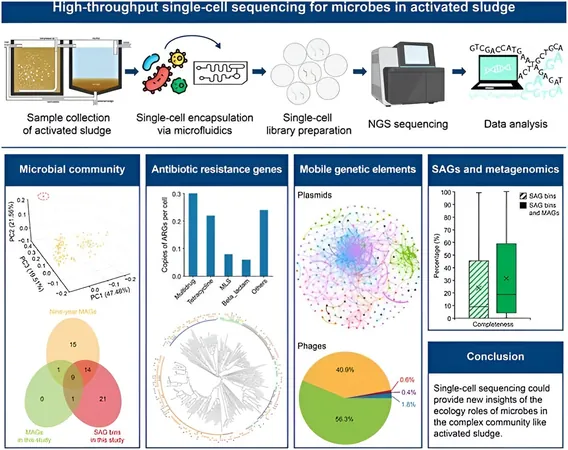
Carried out by a team from the University of Hong Kong and published on September 19, 2024, in the journal Environmental Science and Ecotechnology, this research is set to change the landscape of environmental microbiology. By applying advanced single-cell sequencing to samples collected from a bustling wastewater treatment plant in Hong Kong, the researchers have successfully mapped the genetic networks amongst these microorganisms.
Key Findings
The impressive results of this study included the sequencing of 15,110 individual microbial cells, which were categorized into a staggering 2,454 single-amplified genome bins. Notably, a remarkable 27.5% of these microorganisms were identified as previously unclassified species. The work revealed an astonishing 1,137 antibiotic resistance genes (ARGs), alongside a wealth of plasmids and phages, emphasizing the rampant horizontal gene transfer between species—a phenomenon that underscores urgent public health concerns.
The significance of plasmids in facilitating the dissemination of these ARGs cannot be overstated. This alarming finding highlights an urgent need for enhanced monitoring of wastewater systems to curb the escalating threat of antibiotic resistance, which poses severe risks to public health.
Significance of the Study
This pioneering study marks the first application of high-throughput single-cell sequencing techniques to investigate the activated sludge microbiome specifically. Researchers detected an array of 1,137 ARGs, an impressive 10,450 plasmid fragments, and 1,343 phage contigs, underscoring the dynamic nature of microbial interactions within wastewater. The discovery of shared relationships for plasmid and phage hosts points to a high frequency of gene transfer among microorganisms, heightening the complexity of antibiotic resistance mechanisms in these environments.
Expert Insights
Professor Tong Zhang, the lead researcher, stated, 'Our study showcases the unmatched capabilities of single-cell sequencing in unearthing the concealed dynamics of wastewater treatment systems. The insights gained regarding antibiotic resistance gene transfer are critical in developing better monitoring practices and mitigation strategies, which are essential for protecting public health in line with a One Health framework.'
Broader Implications
The broader implications of this study are profound, particularly in improving wastewater treatment methodologies and controlling the spread of antibiotic-resistant bacteria. As the capability to track ARGs at a single-cell level emerges, it could influence future environmental policies and public health initiatives significantly. Additionally, this groundbreaking technique could pave the way for exploring microbial communities in various ecosystems, including our soils and the intricate microbiomes of the human gut.
Conclusion
In summary, the advent of high-throughput single-cell sequencing not only sheds light on the hidden depths of microbial diversity but also poses essential questions about our health and the environments we inhabit. Stay tuned—this research could change everything we know about antibiotic resistance and its management in the battle for public health safety!



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)