Revolutionizing Microbe Identification: The Game-Changing Nucleotide Database
2025-04-10
Author: Nur
Unlocking the Secrets of Microbial Life
Imagine trying to catalog a library with millions of books where some are misfiled and others authored by the wrong writers. This daunting challenge mirrors the difficulties faced by researchers at Lawrence Livermore National Laboratory (LLNL) as they grappled with the National Center for Biotechnology Information's (NCBI) Nucleotide (nt) database, a colossal repository of DNA sequences from countless species.
The Problem: Outdated and Contaminated Data
The NCBI nt database has transformative potential, aiding in everything from disease diagnosis to environmental monitoring. However, it's become increasingly unwieldy, cluttered with errors and outdated information, according to Nicholas Be, leader of LLNL's Microbiology/Immunology Group. Notably, the database hasn't received updates compatible with the widely-used Centrifuge tool since 2018, hindering accurate DNA classification.
Furthermore, significant contamination issues—inaccurately labeled genetic sequences—threaten to misguide researchers. This contamination can lead to mistaken pathogen identification, potentially derailing scientific studies.
A Groundbreaking Solution
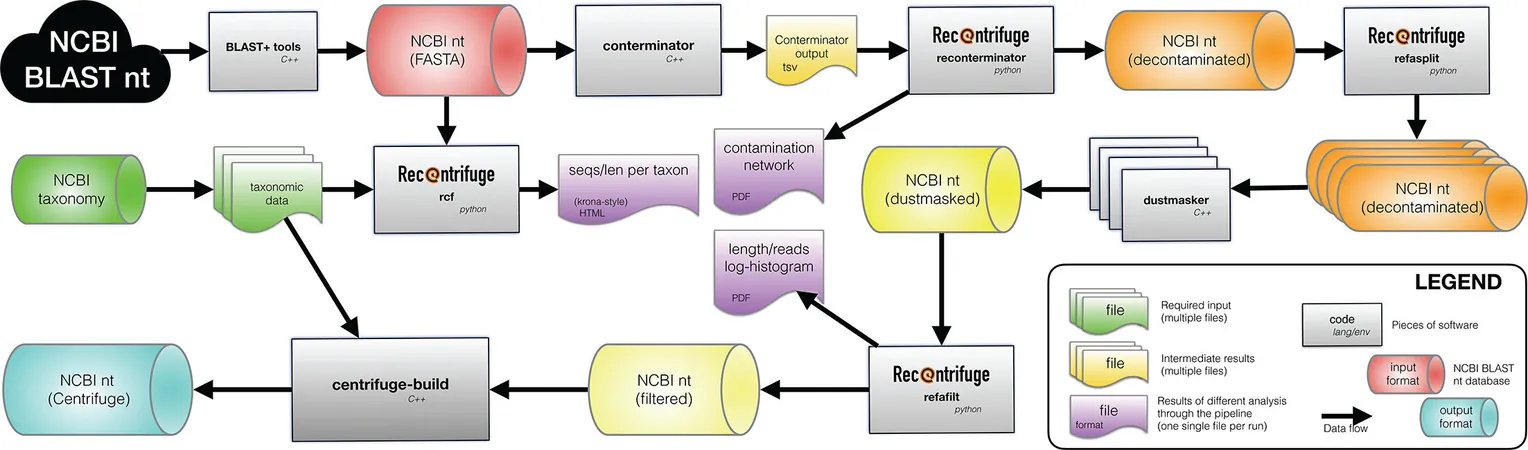
In a pioneering study published in mSystems, LLNL researchers revealed a groundbreaking solution: creating optimized indexes of the nt database that enhance how scientists classify microorganisms. By employing advanced computing technologies, they curated cleaner datasets tailored for Centrifuge, streamlining the process of identifying diverse microorganisms from environmental and clinical samples.
Be stated, "Our new nucleotide database drastically enhances classification accuracy and reliability, allowing for better understanding of the microbial world, irrespective of sample type." This new cleanness in data alignment means fewer errors, particularly critical for understanding malaria-causing Plasmodium species.
Revolutionizing Analysis with Rigorous Quality Control
The team prioritized rigorous quality control to eliminate contaminants and bolster classification accuracy. Their efforts translated into significantly fewer misleading classifications, essential for research in fields ranging from medicine to environmental science.
Broad Applications Across Disciplines
Accurate microbial identification is vital: in medicine, it informs treatment; in environmental science, it assesses ecosystem health; and in forensics, it can determine criminal activity. The researchers hope this newly refined database will raise awareness about the computational resources necessary for maintaining these critical tools.
The Future of Microbial Databases
The LLNL team plans to treat this database as a living resource, akin to software that requires regular updates. With genomic data expanding exponentially, continuous updates are crucial for serving diverse scientific communities. As Jose Manuel Marti, lead author on the project, noted, their transitions to more complex databases reflect a commitment to enhancing scientific accuracy.
Overcoming Computational Hurdles
However, the immense computational demands of updating these databases pose challenges. The initial indexing took over five years of CPU time to complete on a single core. Yet, leveraging parallel processing brought this down significantly, underscoring the necessity of LLNL's contribution to the field.
Accessible Resources for Researchers
Researchers eager to utilize this advancement can access the new decontaminated databases on Amazon Web Services, empowering accurate metagenomic analyses. As efforts continue, the team is now focused on expanding their methodology for different classification engines and tackling specialized databases for viral or fungal identification.
Innovating for the Future
To manage the ever-increasing data deluge, the team is exploring innovative solutions like distributed computing. By breaking classification tasks into simpler components, they aim to further optimize microbe identification efforts for the scientific community.
In summary, LLNL's refreshed nucleotide database not only improves microbial identification but sets a new standard for data integrity in genomics, ensuring that researchers have the most accurate resources available to unearth the hidden complexities of our microbial world.
 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)