Unlocking Cancer's Secrets: How Genetic Collisions Fuel Aggressive Tumors
2024-11-19
Author: Arjun
Introduction
Cancer researchers from the University of Chicago and the University of California, San Francisco (UCSF) have made groundbreaking discoveries that could transform our understanding of aggressive cancers. Their recent study shows that mutations in key genes can lead to severe DNA errors, particularly a type of genetic alteration known as large tandem duplications (TDs). These duplications arise from a collision between two essential cellular processes: transcription and DNA replication.
This new insight could pave the way for innovative treatments aimed at specific cancers exhibiting these genetic changes, as detailed in the November 2024 issue of Nature Cancer.
DNA and Its Integrity
DNA is fundamental to cellular function, serving as the blueprint for life. It must remain reliable and error-free during cell division. The integrity of DNA replication hinges on the stability of the genome, which is supported by intricate DNA repair mechanisms designed to rectify replication errors. However, when these repair systems fail, mutations accumulate, leading to genomic instability—a key factor in cancer development. The specific causes behind these failures have remained a mystery until now.
The Collision of Transcription and Replication
DNA consists of two intertwined strands, functioning both as blueprints for replication and as instructions for protein synthesis—a process known as transcription. Typically, transcription and replication operate in harmony from a safe distance on the DNA molecule. However, they can occasionally clash during transcription and replication collisions (TRCs), leading to disruptions that cause significant cellular stress. The impact of TRCs on genomic instability in human cancers is an area that has yet to be thoroughly investigated.
Research Findings
In their quest to understand the implications of TRCs, Dr. Lixing Yang, an Assistant Professor in the Ben May Department of Cancer Research at the University of Chicago, collaborated with UCSF researchers to identify tumors exhibiting elevated levels of TRCs. By analyzing thousands of tumor samples through whole-genome sequencing, they mapped out various structural variations, including nucleotide deletions, duplications, and translocations.
"Every tumor showcases a unique mix of structural variations, often arising in distinct subsets due to various causal factors," explained Yang. For instance, lung cancers are frequently tied to tobacco use, while melanoma is often prompted by UV exposure. These mutations have unique signatures, and the challenge lies in deciphering them, akin to solving a puzzle made more complex by numerous contributing influences.
Impact of TRCs
The researchers examined data from an impressive 6,193 whole-genome-sequenced tumors to assess how TRCs contribute to genomic instability. They discovered that structural variations resulting from these collisions exhibit a telling signature that can be identified through dosage imbalances—variations in the number of DNA copies at junctions where strands converge.
This DNA 'patching' phenomenon occurs when excess copies of a strand mistakenly attach to other regions, leading to structural variations. In many cases, this linking happens near the original sequence, establishing repetitive patterns known as tandem duplications.
Key Conclusions
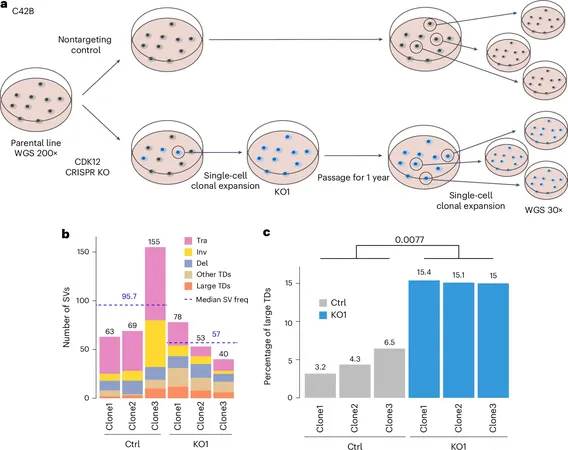
The exciting conclusion of this study? Large TDs have a higher prevalence in cancers of the upper gastrointestinal tract, prostate cancer, and in female-related cancers like breast and ovarian cancer. Alarmingly, these large TDs are associated with poor patient survival rates and correlate strongly with mutations in critical genes such as TP53, CDK12, and SPOP.
While the correlation had been acknowledged, this research definitively links the genetic collisions to the CDK12 mutations that induce large TDs—a revelation that opens new pathways for targeted therapies.
Future Implications
The study underscores that cancers marked by large TDs are particularly responsive to certain drugs, including WEE1, CHK1, and ATR inhibitors. This presents a promising avenue for treatment options for those battling aggressive cancers driven by these genetic alterations.
As we delve deeper into the molecular mechanisms behind these disease processes, hope glimmers for more effective strategies that could vastly improve the prognosis and quality of life for cancer patients facing difficult battles against it. Stay tuned as we continue to uncover the complex relationships between genetics and cancer!

 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)