Unraveling the Influenza A Virus: A Comprehensive Analysis of Genomic Surveillance in Sri Lanka
2024-11-26
Author: John Tan
Introduction
Influenza A continues to be a global health threat, recognized by the World Health Organization (WHO) as a priority pathogen due to its pandemic potential. Understanding the genetic makeup and evolution of influenza strains is crucial for timely vaccine development and implementation, particularly in regions like Sri Lanka, which experiences seasonal outbreaks. Our study delves into the molecular epidemiology of influenza A in Sri Lanka's Western Province and emphasizes the need for ongoing genomic surveillance.
Study Overview
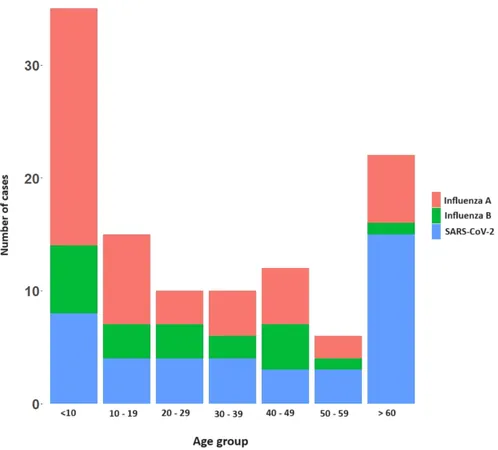
Between November 2022 and May 2024, our research team recruited 349 individuals exhibiting fever and respiratory symptoms from two major hospitals. Specimens were collected and subjected to quantitative PCR screening for Influenza A, Influenza B, and SARS-CoV-2. Subsequent genomic sequencing was performed on the identified influenza A virus strains using advanced Oxford Nanopore Technology.
Key Findings
Our results revealed that Influenza A was present in 14% of the participants, while Influenza B accounted for 5.7%, and SARS-CoV-2 was detected in 11.7%. Co-infections were noted in five individuals, with four demonstrating co-infection of influenza A and B, and one with both influenza A and SARS-CoV-2. Noteworthy was the phylogenetic analysis which classified H1N1 HA gene sequences into subclades C.1, C.1.2, and C.1.8 for the year 2023, and C.1.8 and C.1.9 for 2024. The H3N2 analysis revealed a shift from the 3 C.2a1b.2a.2a.1b clade in 2023 to the 3 C.2a1b.2a.2a.3a.1 clade in 2024. The presence of various amino acid substitutions in the H1N1 and H3N2 strains indicates significant evolutionary changes, warranting further investigation into their potential impact on vaccine efficacy.
Implications for Public Health
Sri Lanka, a tropical country, faces unique challenges regarding influenza outbreaks, which tend to peak during the rainy seasons. With an estimated 3.2 million cases of severe influenza occurring globally each year, the WHO emphasizes the importance of effective vaccination strategies tailored to local virus dynamics. Our findings underscore the necessity for a flexible, evidence-based vaccination approach, taking into account local epidemiological data. The fluctuation of dominant strains observed in our study, particularly the rise and fall of H1N1 and H3N2 subtypes, highlights the importance of real-time surveillance. As the virus evolves, genomic analysis will be critical for informing vaccine formulation, enhancing community resilience against seasonal outbreaks.
Looking Ahead: The Future of Influenza Surveillance
Our study serves as a vital first step toward establishing a comprehensive genomic surveillance system in Sri Lanka. However, the relatively small sample size limits the generalizability of the findings. Nationwide surveillance efforts incorporating functional assays will be crucial in understanding the biological significance of detected mutations, as well as in tailoring public health responses. Given the unpredictable nature of viral evolution, the WHO recommends an adaptive vaccination strategy that accounts for local strains and their behavior. Continuous monitoring will not only assist in effectively scheduling vaccination campaigns but will also help to mitigate the impact of the next potential pandemic.
Conclusion
In conclusion, understanding the genetic landscape of circulating influenza A strains in Sri Lanka over the past 18 months provides invaluable insights for vaccine selection and public health strategies. The genetic diversity observed, coupled with certain mutations of indeterminate significance, presents both challenges and opportunities for future research. Ongoing genomic surveillance is essential for keeping pace with viral changes, ensuring that public health interventions remain effective against influenza and associated threats. As we move forward, it’s crucial for health authorities to integrate these findings into robust public health frameworks to better protect the population. Stay tuned for more updates as we continue our vital research on infectious diseases in Sri Lanka!
 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)