Revolutionary Low-Cost Phenotyping Technology Reveals Breakthroughs in Disease-Resistant Crop Research!
2024-11-20
Author: Nur
Introduction
In an exciting advancement for agricultural science, researchers have leveraged a low-cost phenotyping system to unlock critical insights into the genetic diversity of Quantitative Disease Resistance (QDR) in wild tomato species. This innovative approach could pave the way for breeding more resilient crops capable of withstanding various plant diseases.
Understanding Quantitative Disease Resistance (QDR)
Quantitative Disease Resistance, or QDR, represents a robust form of plant defense that offers durable, partial protection against a wide range of pathogens. Unlike qualitative resistance, which is primarily governed by single resistance (R) genes, QDR is polygenic, meaning it’s influenced by multiple genes and can manifest in various ways. This includes delayed onset of symptoms and reduced susceptibility to infections—characteristics that are crucial for crop survival, especially in an era of increasing plant diseases and environmental challenges.
The Landmark Study
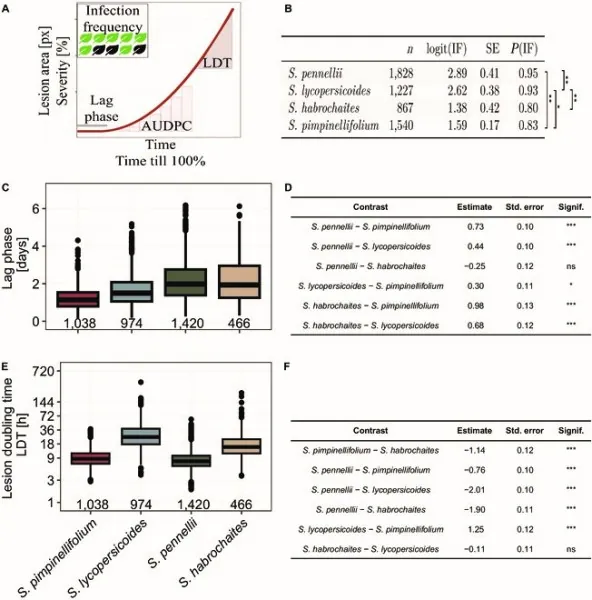
A landmark study published on August 5, 2024, in *Plant Phenomics* outlines the remarkable findings from a research team that investigated QDR in four wild tomato species: *Solanum habrochaites*, *S. lycopersicoides*, *S. pennellii*, and *S. pimpinellifolium*. The team applied the revolutionary 'Navautron' automated phenotyping system, which took continuous images of infected leaves and employed advanced algorithms to analyze vital parameters related to disease progression.
Key Findings from the Research
The results were illuminating. The analysis highlighted significant phenotypic diversity in QDR among the species studied. Notably, *S. pimpinellifolium* displayed the shortest lag phase before lesions formed—a mere 36.2 hours—while *S. habrochaites* and *S. pennellii* showed longer lag phases of around 59 hours. Furthermore, the study revealed that lesion growth rates varied widely: *S. pimpinellifolium* and *S. pennellii* exhibited rapid lesion expansion with doubling times of just 11 hours, whereas *S. habrochaites* and *S. lycopersicoides* were significantly slower, with doubling times of 36 and 41 hours, respectively.
Intraspecific Variation and Superior Accessions
In addition to infection frequency, which varied with *S. habrochaites* showing the lowest rate at 80%, the researchers discovered substantial intraspecific variation within *S. pennellii*. This species demonstrated a broad spectrum of responses to disease, indicating that some accessions are genetically superior in fighting infections.
Expert Insights
According to Dr. Remco Stam, the lead researcher of the study, "Unlocking the potential of QDR in crop breeding has been a long-standing challenge. Our study showcases a cost-effective phenotyping system that can provide high-resolution data crucial for understanding and utilizing QDR traits in wild crop relatives."
Implications for Sustainable Agriculture
The implications of this research extend far beyond tomato cultivation. The ability to dissect and understand the intricate mechanisms behind QDR offers a beacon of hope for sustainable agriculture. As climate change and various pathogens threaten global food security, insights from this study may lead to the development of crops that can naturally resist diseases without heavy dependence on chemical fungicides or singular resistance genes.
Conclusion
As we move forward, the integration of such advanced but affordable technology could revolutionize not just tomato breeding but also agricultural practices, fostering healthier ecosystems and ensuring that our crops can thrive in increasingly stressful environments.
Stay Tuned!
Stay tuned to learn how this groundbreaking research continues to change the landscape of agriculture and plant resilience!
 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)